Expanding the Genetic Code
Expanding the genetic code - beyond the 20 natural amino acids
All known life uses a limited set of 20-22 amino acids to synthesize proteins. In recent years it has become possible to genetically encode an expanded set of designer amino acids with tailored chemical and physical properties into proteins in bacteria and eukaryotes by reprogramming the genetic code and rewiring the translational machinery.
These strategies have started to have a big impact on biological studies, as they enable diverse applications, including approaches for imaging and probing proteins, controlling and manipulating protein activity as well as engineering and designing new protein functions and protein therapeutics.
In the KLang lab we are developing and advancing approaches to expand the genetic code and endow proteins with novel chemical reactivities within their physiological environment. To extend the chemical space and functional repertoire of proteins withing living cells, we use rational and evolutionary approaches aimed at engineering components of the translational machinery that enable the site-specific incorporation of custom-designed non-canonical amino acids (ncAAs) into proteins in living cells.
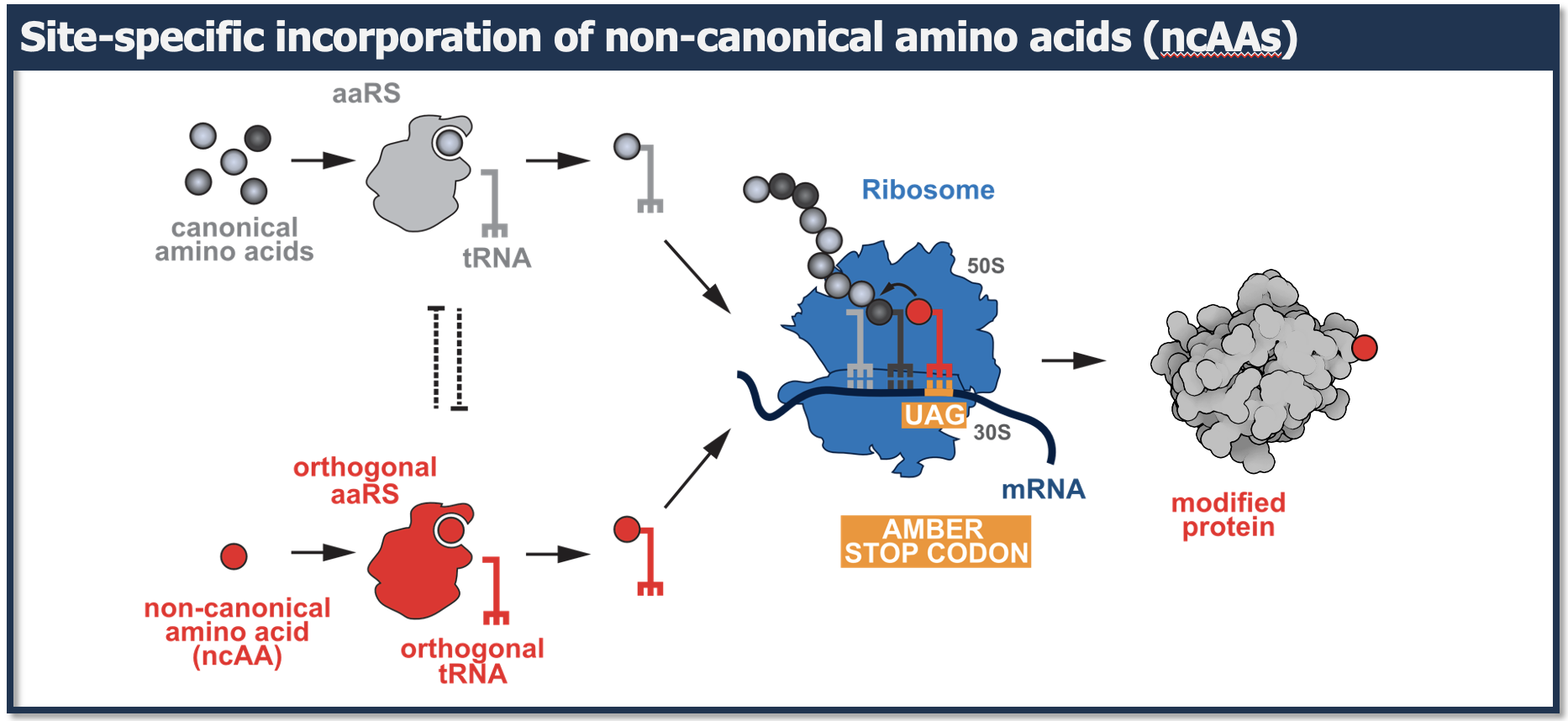
By equipping proteins with functionalities beyond the ones provided by the 20 natural amino acids we aim to:
- design and apply bioorthogonal reactions to image and label proteins
- develop chemical tools to validate and manipulate protein-protein interactions
- deploy novel chemoenzymatic approaches to study ubiquitylation/deubiquitylation pathways