Mimicking Post-Translational Modifications
Novel tools to decipher the ubiquitin code
In a third research line we are pioneering approaches where site-specifically introduced ncAAs serve as a platform for tailored, non-endogenous chemoenzymatic reactions. We have developed approaches to site-specifically ubiquitylate target proteins both in vitro and in vivo.
Post-translational modification of proteins with ubiquitin (Ub) and Ub-like modifiers (Ubls) is central to the regulation of eukaryotic cellular processes. Substrate modifications range from a single Ub moiety being attached to a target protein to complex polymeric Ub/Ubl chains with distinct topologies that control the activity, stability, interaction and localization of almost all cellular proteins and elicit a variety of biological outputs. Our ability to characterize the roles of distinct Ub/Ubl patterns and to identify enzymes and receptors that create, recognize and remove these modifications is however hampered by the difficulty to prepare them.
Our lab develops tools to study aspects of ubiquitylation that are challenging or impossible to be addressed by more traditional technologies. We combine genetic code expansion approaches with transpeptidase engineering to develop toolkits for generating defined Ub/Ubl architectures, including mixed, branched and hybrid Ub/Ubl chains, which allows us to study the functional impact of these complex types of modifications on critical cellular processes. Furthermore, we develop approaches based on genetic code expansion and novel bioorthogonal and crosslinking chemistries to profile activities and specificities of the enzymes that install, recognize and revert Ub/Ubl modifications.
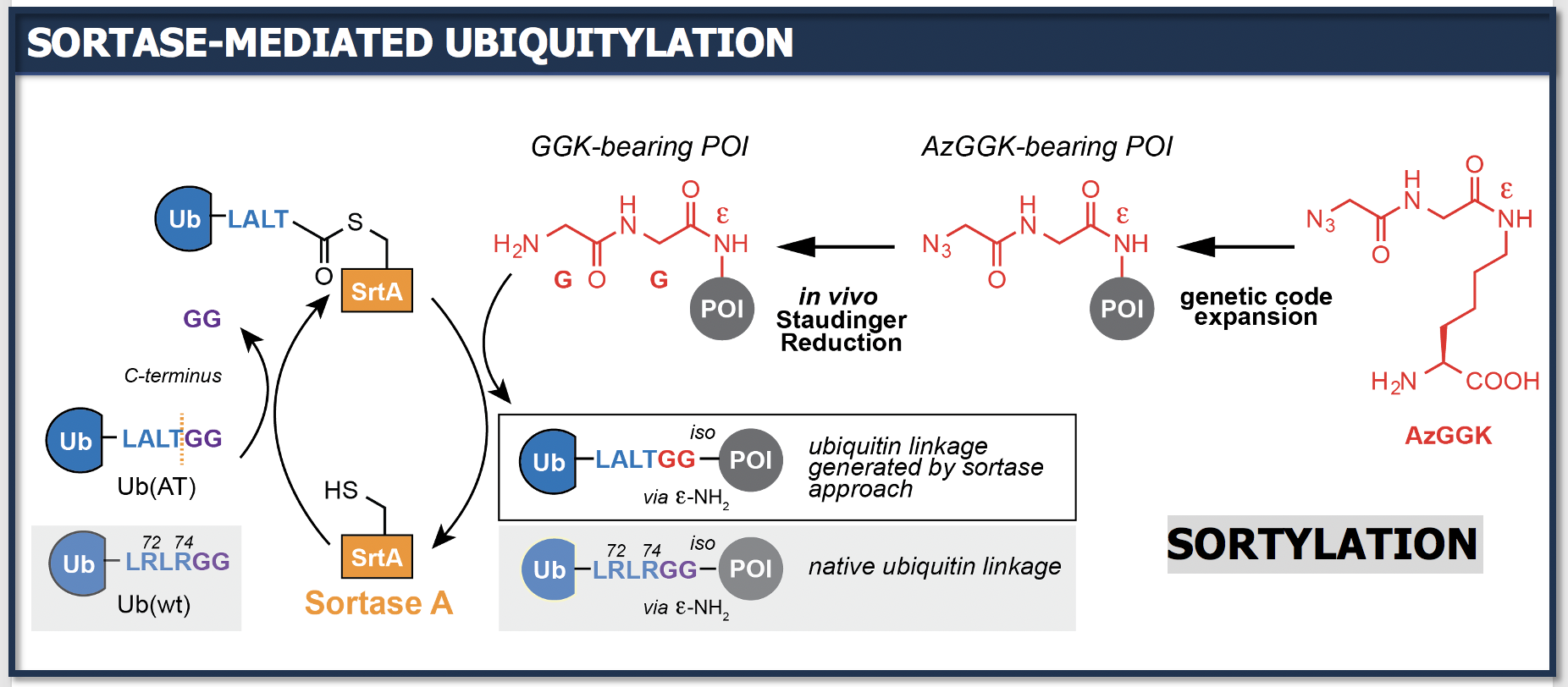
Selected publications:
- M. Fottner, M. Weyh, S. Gaussmann, D. Schwarz, M. Sattler, K. Lang*; A modular toolbox to generate complex polymeric ubiquitin architectures using orthogonal sortase enzymes; Nat. commun. 2021, 6515, 12
- M. Fottner, A.-D. Brunner, V. Bittl, D. Horn-Ghetko, A. Jussupow, V. RI Kaila, A. Bremm, K. Lang*; Site-specific ubiquitylation and SUMOylation using genetic-code expansion and sortase; Nat. Chem. Biol. 2019, 15, 276